Identifying the target affinities of various tautomeric isoforms during drug discovery is vital for optimizing a drug's efficacy, safety, and pharmacokinetic properties. This is because different tautomers of a molecule can exhibit varied biological activities, stabilities, and interactions with the target.
Free Energy Perturbation (FEP) calculations can be used to predict favored tautomeric forms inside the target binding pocket, but running these calculations for each of these tautomers separately is tedious and time-inefficient. Luckily, you can accurately characterizeall tautomeric forms at once in a single TandemFEP run.
Running tautomeric isomers in TandemFEP is easy. You can create a dataset from multiple sketches - one for each tautomer
In this demo, let’s see how you can run multiple tautomeric isomers at once in TandemFEP using edaravone as an example. This FDA-approved drug for the treatment of amyotrophic lateral sclerosis (ALS) exists in three forms of tautomeric isomers - keto, enol, and amine.
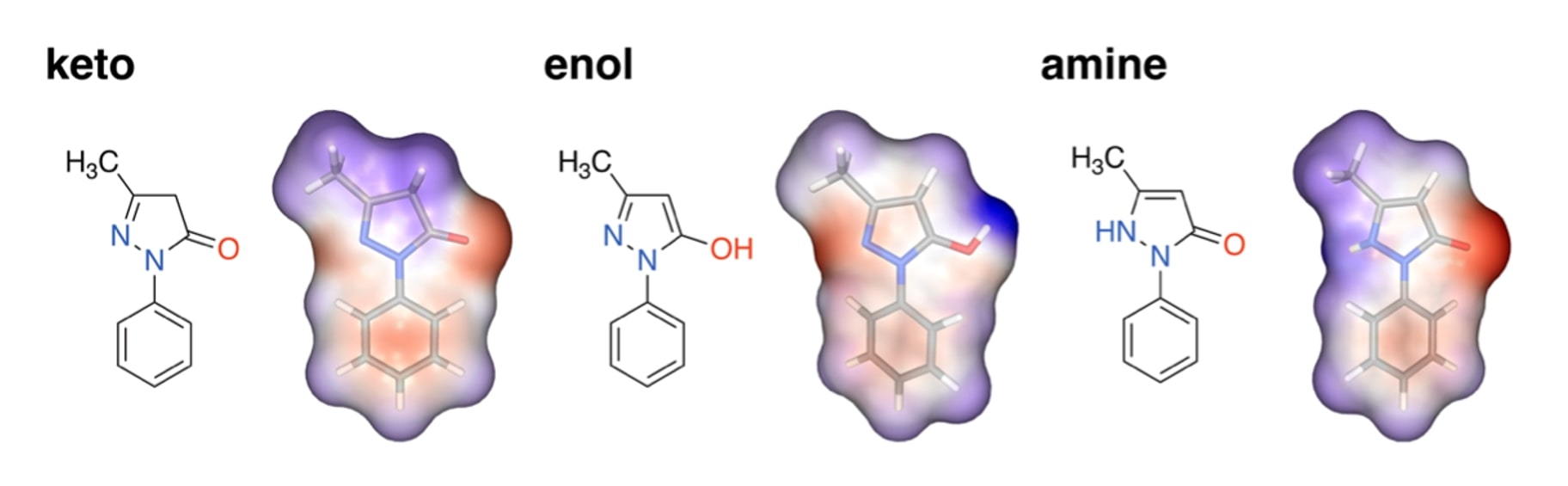
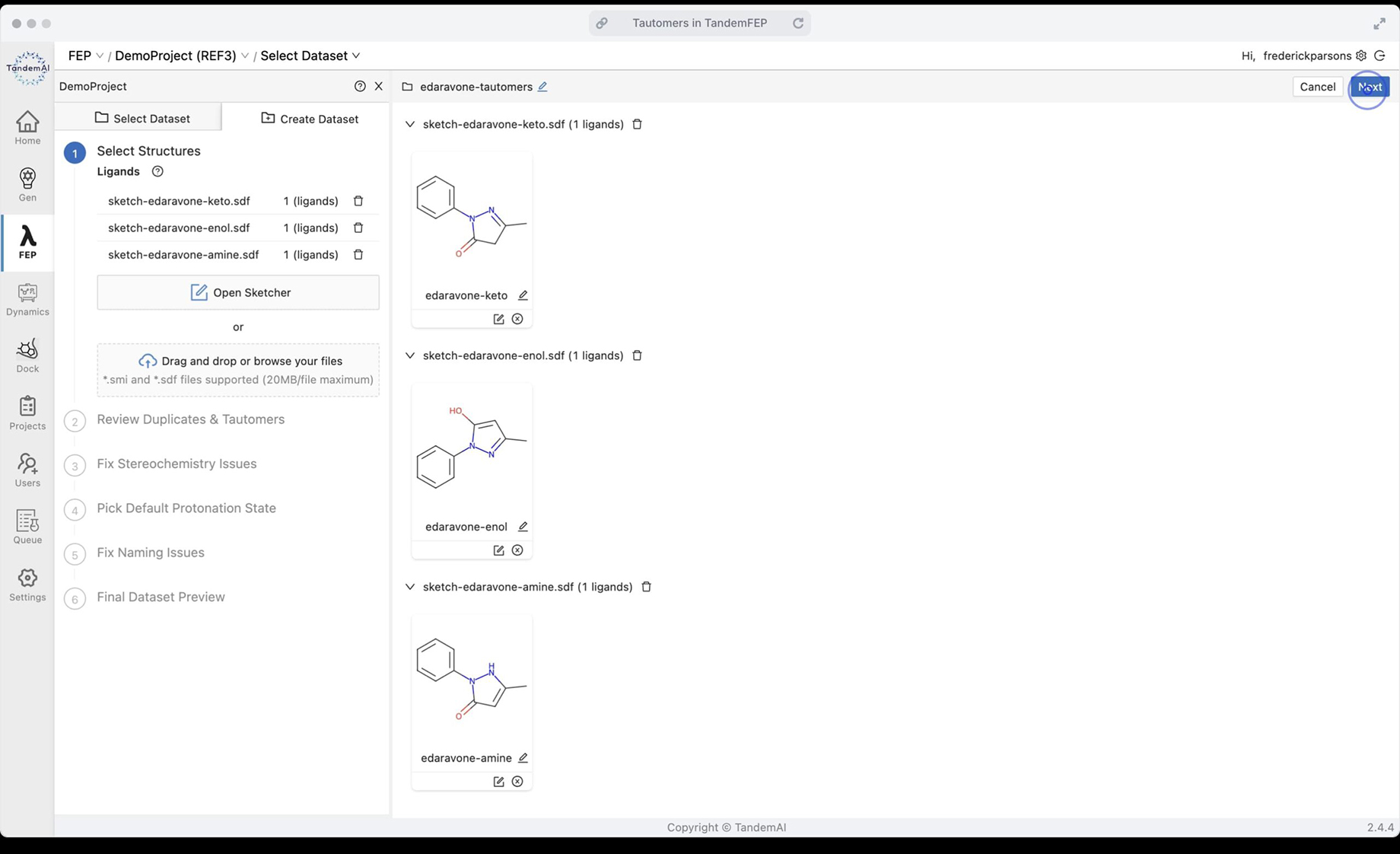
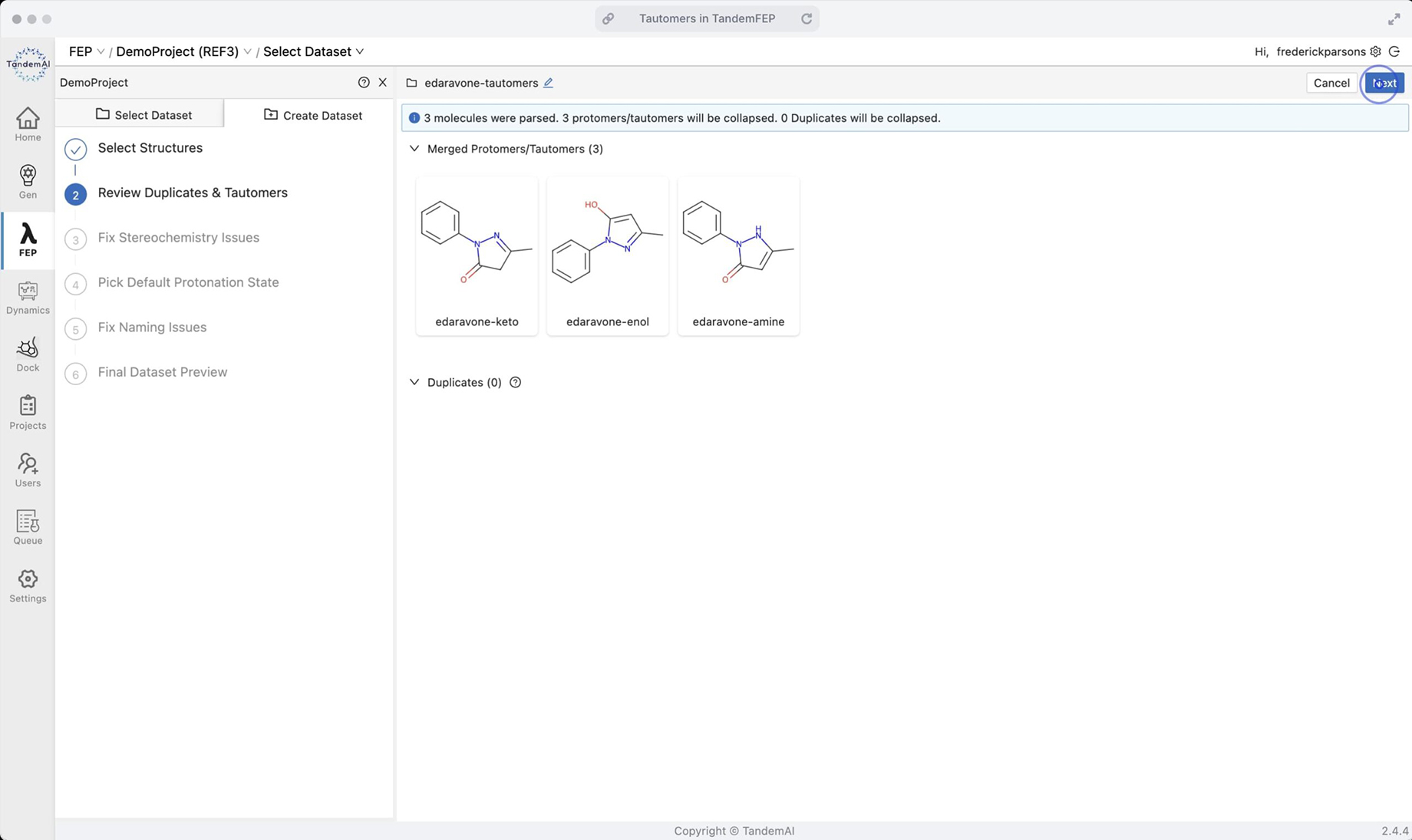
TandemViz automatically detects that these three compounds are tautomers. In the dataset, they are collapsed into one representative parent molecule.
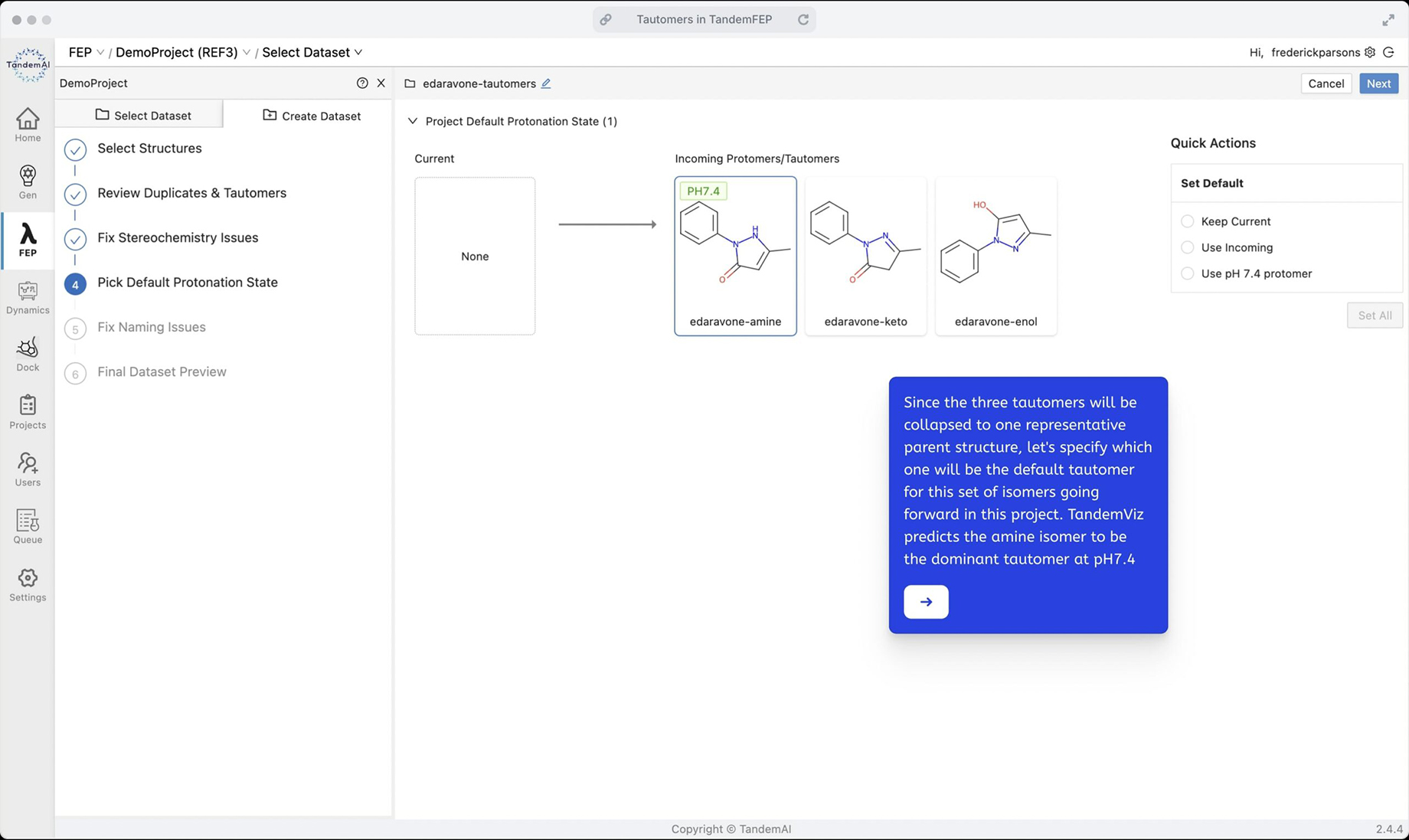
Next, choose your default tautomer for this set. In this case, TandemViz determined that the amine is the most probable tautomer at pH 7.4, so use it as the default tautomer.
Open the dataset in TandemFEP to see it display only the default tautomer. For this experiment, specify that you want to run TandemFEP for all three tautomers.
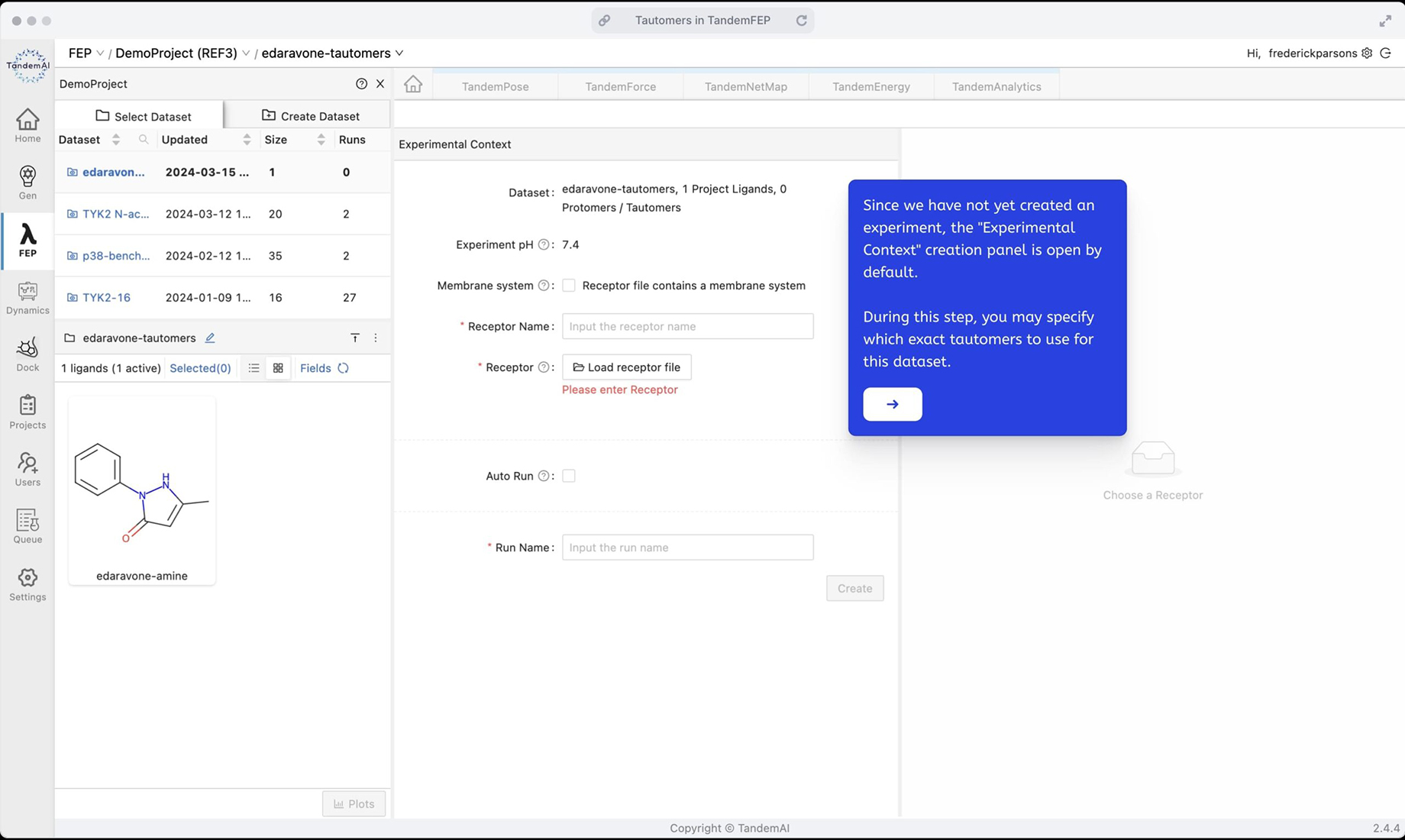
Specify the set of tautomers for this experiment context using the dataset pane option to upload an SDF file containing the three desired tautomers.
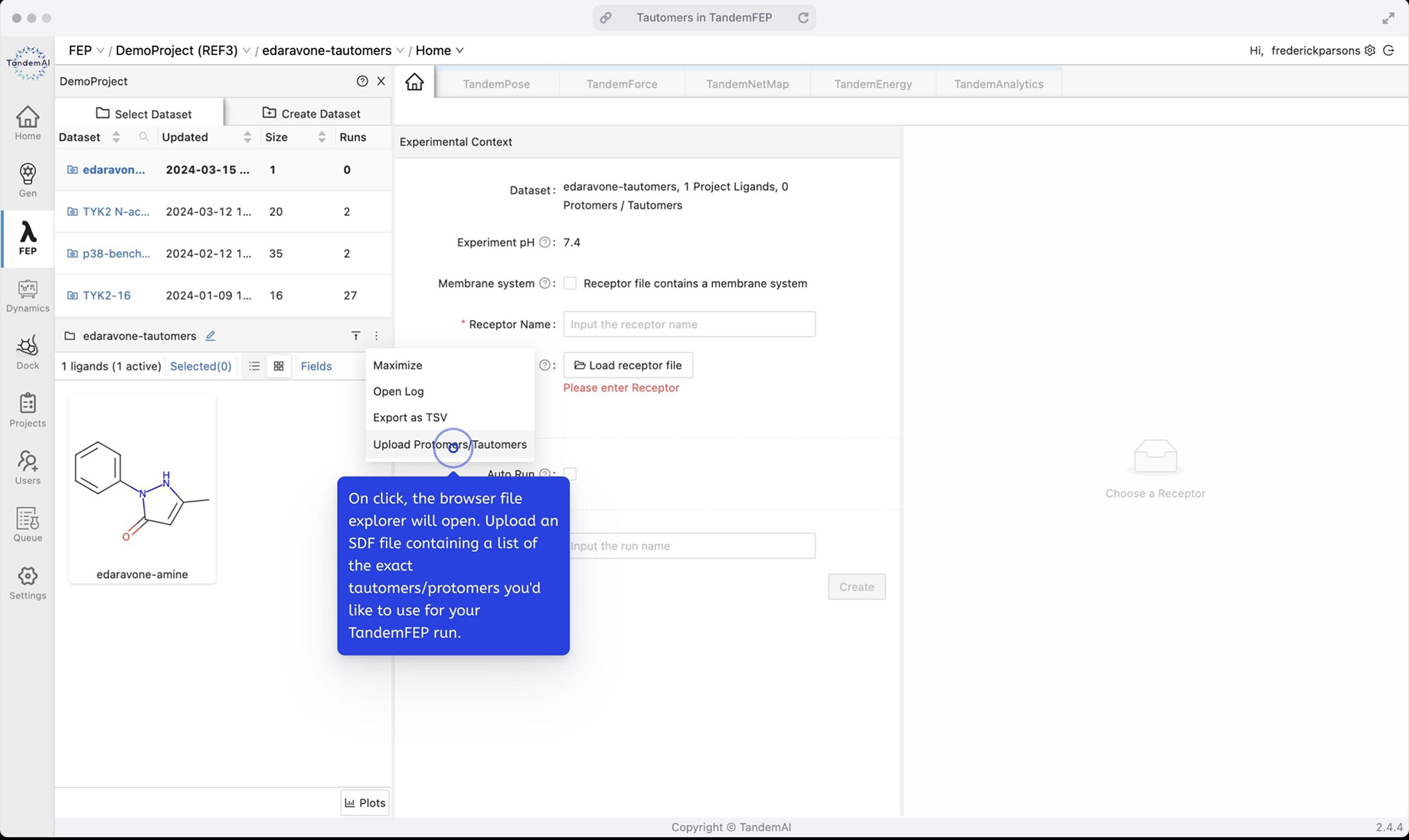
On upload, the dataset pane refreshes with the list of tautomers specified for each parent ligand in the dataset – in this example case, there was just one. You can now see it expanded to three tautomers.
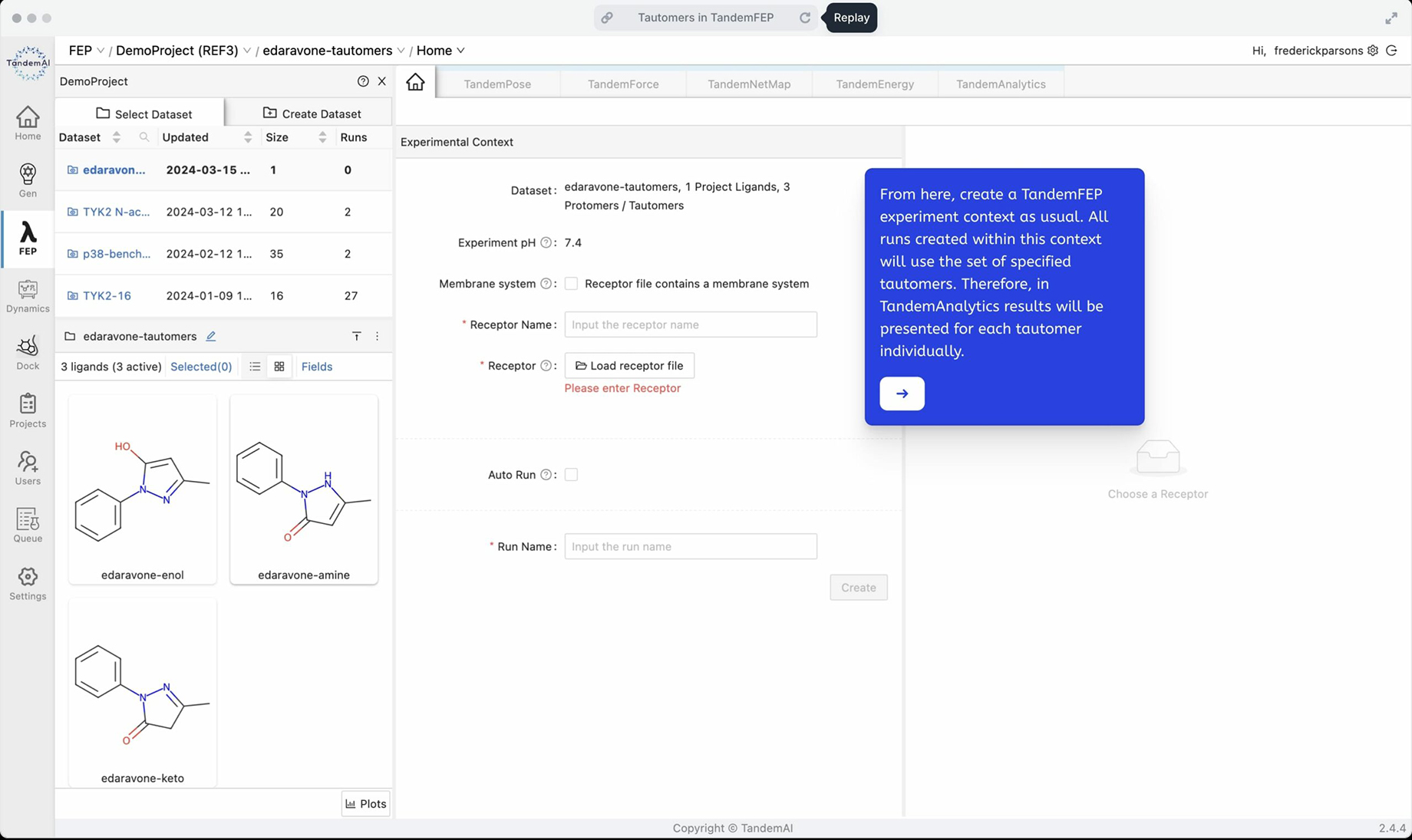
From here, proceed as normal and create the TandemFEP experiment context. All runs contained within this context will be for the three specified tautomers, saving you the time it would take to run each of the three separately. TandemFEP is versatile in handling your specific case, and this tutorial also applies to molecules with 2 tautomers or more than 3 tautomers.
If you’re still having trouble, click here for an interactive tutorial to walk you through the process.